OLS
> fit.ols<-glm(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 + lmhval +
+ phnew + phisp, data = philly2)
> summary(fit.ols)
Call:
glm(formula = usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac +
ph70 + lmhval + phnew + phisp, data = philly2)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 534.491 164.270 3.254 0.00124 **
lmhhinc 2.462 12.176 0.202 0.83990
lpop -1.344 6.338 -0.212 0.83216
pnhblk 21.158 18.077 1.170 0.24260
punemp -5.097 63.645 -0.080 0.93622
pvac 371.699 58.427 6.362 5.96e-10 ***
ph70 -79.691 35.535 -2.243 0.02552 *
lmhval -45.668 10.458 -4.367 1.64e-05 ***
phnew 17.958 319.042 0.056 0.95514
phisp -56.308 30.695 -1.834 0.06741 .
—
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 4829.927)
Null deviance: 2938287 on 375 degrees of freedom
Residual deviance: 1767753 on 366 degrees of freedom
AIC: 4268.4
Number of Fisher Scoring iterations: 2
GWR
gwr.fit1<-gwr(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 + lmhval +phnew + phisp, data = philly2.sp, bandwidth = gwr.b1, se.fit=T, hatmatrix=T)
> gwr.fit1
Call:
gwr(formula = usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac +
ph70 + lmhval + phnew + phisp, data = philly2.sp, bandwidth = gwr.b1,
hatmatrix = T, se.fit = T)
Kernel function: gwr.Gauss
Fixed bandwidth: 1322.708
Summary of GWR coefficient estimates at data points:
Min. 1st Qu. Median 3rd Qu. Max. Global
X.Intercept. -1574.4098 -53.8875 88.4952 472.7282 3092.1466 534.4908
lmhhinc -151.0306 -7.0538 3.2205 22.3099 120.2753 2.4616
lpop -76.6700 1.1576 7.2067 20.4788 109.5747 -1.3441
pnhblk -124.9781 -2.0948 44.5163 100.0885 490.8730 21.1576
punemp -627.4200 -150.5909 -17.8892 69.6271 752.1507 -5.0966
pvac -1329.2458 2.5473 165.4452 343.9353 1108.9034 371.6993
ph70 -1028.8902 -161.7810 -43.4011 -8.2491 144.6265 -79.6910
lmhval -178.5925 -70.3725 -26.7389 -3.7657 89.1748 -45.6676
phnew -3747.6137 -484.6544 54.6557 734.6135 6434.5611 17.9575
phisp -313.3416 -24.9975 4.8295 117.2091 1533.6439 -56.3076
Number of data points: 376
Effective number of parameters (residual: 2traceS – traceS’S): 220.8092
Effective degrees of freedom (residual: 2traceS – traceS’S): 155.1908
Sigma (residual: 2traceS – traceS’S): 59.06332
Effective number of parameters (model: traceS): 178.5045
Effective degrees of freedom (model: traceS): 197.4955
Sigma (model: traceS): 52.3567
Sigma (ML): 37.9452
AICc (GWR p. 61, eq 2.33; p. 96, eq. 4.21): 4491.91
AIC (GWR p. 96, eq. 4.22): 3979.926
Residual sum of squares: 541379.3
Quasi-global R2: 0.81575
> gwr.b2<-gwr.sel(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 + lmhval +phnew + phisp, data = philly2.sp, gweight = gwr.bisquare)
> gwr.fit2<-gwr(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 + lmhval +phnew + phisp, data = philly2.sp, bandwidth = gwr.b2, gweight = gwr.bisquare, se.fit=T, hatmatrix=T)
> gwr.fit2
Call:
gwr(formula = usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac +
ph70 + lmhval + phnew + phisp, data = philly2.sp, bandwidth = gwr.b2,
gweight = gwr.bisquare, hatmatrix = T, se.fit = T)
Kernel function: gwr.bisquare
Fixed bandwidth: 5092.898
Summary of GWR coefficient estimates at data points:
Min. 1st Qu. Median 3rd Qu. Max. Global
X.Intercept. -649.3890 -5.7699 134.6249 512.9574 2336.5957 534.4908
lmhhinc -180.3145 -4.4545 1.7487 13.7554 68.2914 2.4616
lpop -49.1608 1.2314 6.3430 19.0823 69.7005 -1.3441
pnhblk -106.4233 1.3658 41.0256 96.5291 285.2134 21.1576
punemp -397.5988 -143.8982 -6.2685 57.4553 729.4700 -5.0966
pvac -757.5534 8.8245 209.8576 370.7793 650.3669 371.6993
ph70 -643.0070 -207.9799 -66.3040 -19.8028 142.9682 -79.6910
lmhval -150.2726 -69.5496 -34.8198 -6.7118 107.7625 -45.6676
phnew -1844.6086 -418.1211 19.6153 509.9117 7421.2055 17.9575
phisp -221.0604 -26.5670 -7.5865 84.2566 1418.3152 -56.3076
Number of data points: 376
Effective number of parameters (residual: 2traceS – traceS’S): 132.4964
Effective degrees of freedom (residual: 2traceS – traceS’S): 243.5036
Sigma (residual: 2traceS – traceS’S): 62.2312
Effective number of parameters (model: traceS): 107.6713
Effective degrees of freedom (model: traceS): 268.3287
Sigma (model: traceS): 59.2826
Sigma (ML): 50.0803
AICc (GWR p. 61, eq 2.33; p. 96, eq. 4.21): 4316.932
AIC (GWR p. 96, eq. 4.22): 4117.761
Residual sum of squares: 943021.6
Quasi-global R2: 0.6790573
gwr.b3<-gwr.sel(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 +
lmhval + phnew + phisp, data = philly2.sp, adapt = TRUE)
gwr.fit3<-gwr(usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac + ph70 + lmhval +
+ phnew + phisp, data = philly2.sp, adapt=gwr.b3, se.fit=T, hatmatrix=T)
> gwr.fit3
Call:
gwr(formula = usarea ~ lmhhinc + lpop + pnhblk + punemp + pvac +
ph70 + lmhval + phnew + phisp, data = philly2.sp, adapt = gwr.b3,
hatmatrix = T, se.fit = T)
Kernel function: gwr.Gauss
Adaptive quantile: 0.02491844 (about 9 of 376 data points)
Summary of GWR coefficient estimates at data points:
Min. 1st Qu. Median 3rd Qu. Max. Global
X.Intercept. -1413.25718 2.04814 150.67770 593.38119 2856.09861 534.4908
lmhhinc -77.30238 -6.62505 2.08877 20.59832 121.03243 2.4616
lpop -71.53993 0.32328 6.55222 19.42020 93.59455 -1.3441
pnhblk -139.33868 -0.35274 39.43998 102.07286 462.87992 21.1576
punemp -592.27650 -109.64202 -3.93096 63.56270 623.38186 -5.0966
pvac -1410.12965 11.95427 193.34738 350.39251 1047.77143 371.6993
ph70 -975.65611 -190.62161 -67.38336 -13.17506 137.47857 -79.6910
lmhval -185.48730 -73.39044 -36.70912 -7.56967 48.91389 -45.6676
phnew -2570.54553 -577.37945 29.21937 654.40082 4045.23829 17.9575
phisp -182.91660 -29.72723 -7.23980 65.71058 771.29484 -56.3076
Number of data points: 376
Effective number of parameters (residual: 2traceS – traceS’S): 177.8408
Effective degrees of freedom (residual: 2traceS – traceS’S): 198.1592
Sigma (residual: 2traceS – traceS’S): 54.21695
Effective number of parameters (model: traceS): 135.2358
Effective degrees of freedom (model: traceS): 240.7642
Sigma (model: traceS): 49.18654
Sigma (ML): 39.35938
AICc (GWR p. 61, eq 2.33; p. 96, eq. 4.21): 4258.02
AIC (GWR p. 96, eq. 4.22): 3964.174
Residual sum of squares: 582484.4
Quasi-global R2: 0.8017605
> gwr.fit1$bandwidth
[1] 1322.708
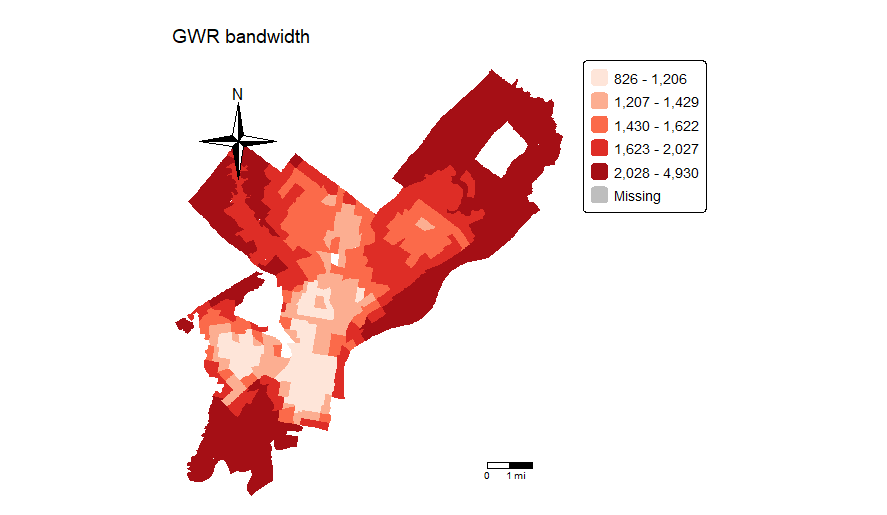
> philly2$bwadapt <- gwr.fit3$bandwidth
> tm_shape(philly2, unit = “mi”) +
+ tm_polygons(col = “bwadapt”, style = “quantile”,palette = “Reds”,
+ border.alpha = 0, title = “”) +
+ tm_scale_bar(breaks = c(0, 1, 2), size = 1, position = c(“right”, “bottom”)) +
+ tm_compass(type = “4star”, position = c(“left”, “top”)) +
+ tm_layout(main.title = “GWR bandwidth”, main.title.size = 0.95, frame = FALSE, legend.outside = TRUE)